Framework to solve problems based on ODE and PDE
C++17 and C++20
Python layer using Pybind11
Seamless parallelism with default communicator including ensemble runs
Powerful interpolation and integration operators working in parallel
Advanced Post-processing including for high order approximation and high order geometry
Build: CMake and CMake Presets
Docs: https://docs.feelpp.org including dynamic pages that can be downloaded as notebooks
DevOps:
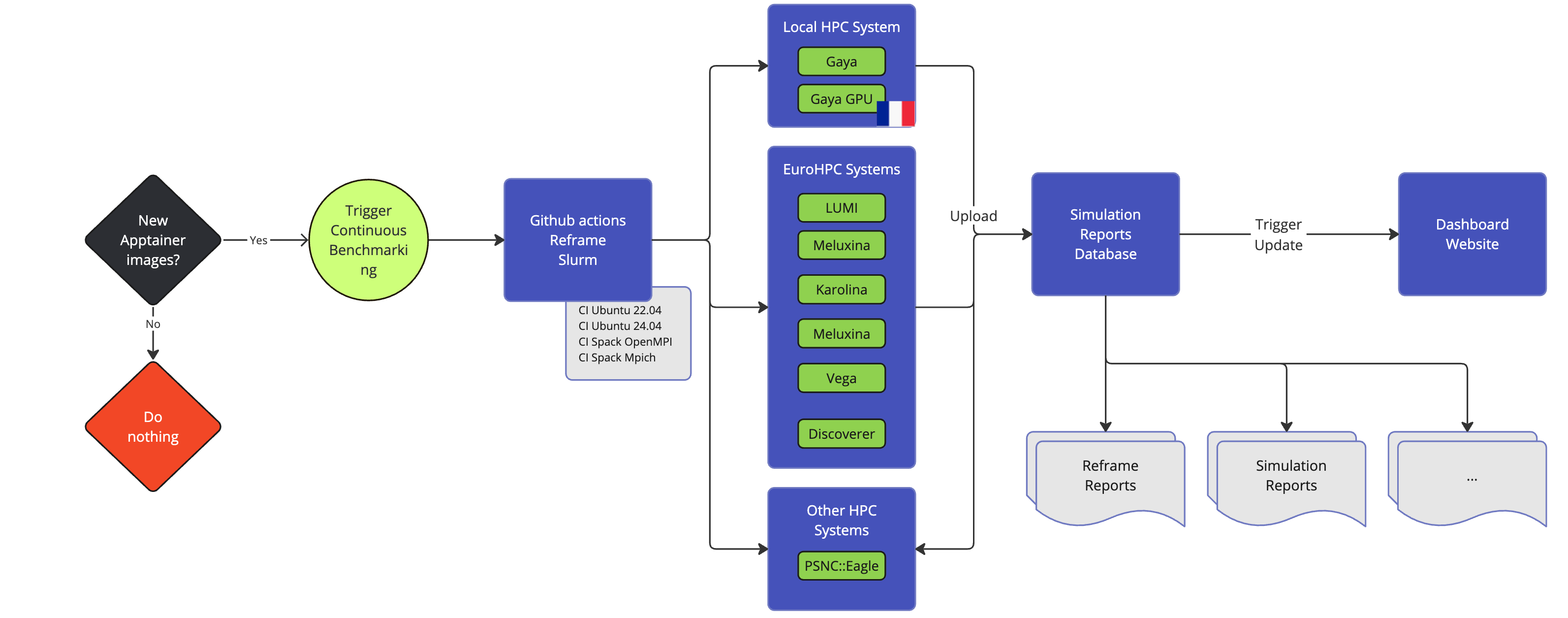
GitHub Actions: CI/CD and Continuous Benmarking on inHouse and EuroHPC systems
Packaging: Ubuntu/Debian, spack, MacPort
Containers: Docker, Apptainer
Tests: About a thousand tests in sequential and parallel C++ and Python
Usage: Research, R&D, Teaching, Services
The Feel++ software
automation, code generation, applications
Christophe Prud'homme -- University of Strasbourg
Cemosis
What is Cemosis ?
Created in 2013, led by C Prud’homme
Hosted by IRMA (Institute for Advanced Mathematical Research) - Unistra
Organized in 3 poles :
Projects C Prud’homme
Training M Maslek
Hardware and software V Chabannes
Labeled Unistra platform in 2021 by the new Cortecs network (cortecs.unistra.fr)
Currently(2024/10) 7 engineers
Cemosis Software Eco-system
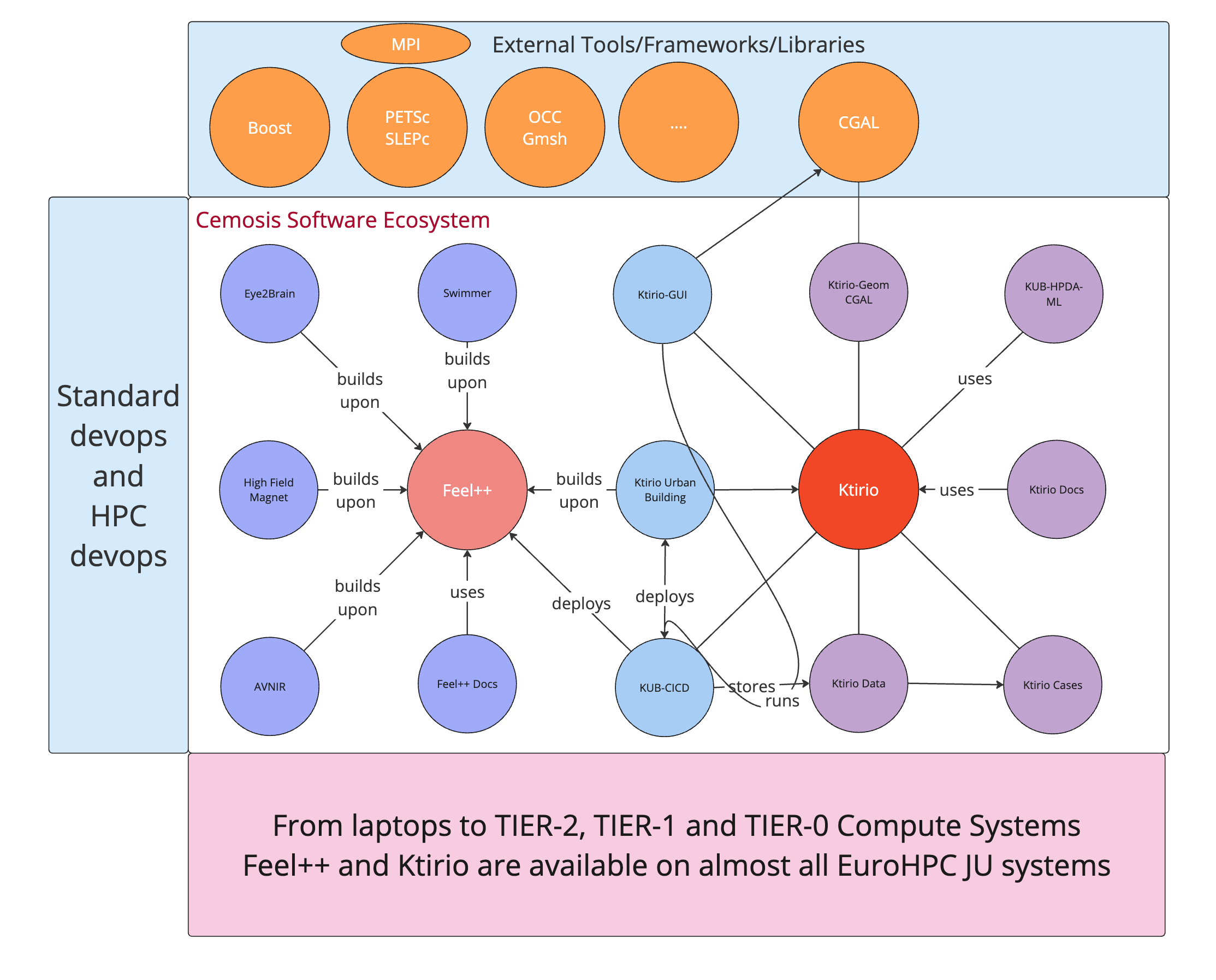
Our Current Big Projects
PEPR NumPEX French Exascale project : Project Exa-MA (Methods and Algorithms at Exascale)
use Citation File Format
Data: Benchmark data input/output and software, example: An Exa-MA benchmark dataset
CoE Hidalgo2 : Urban Building Energy Modeling
use Citation File Format
Data: Mesh generation, models and report results
Feel++ Suite
What is Feel++ ?
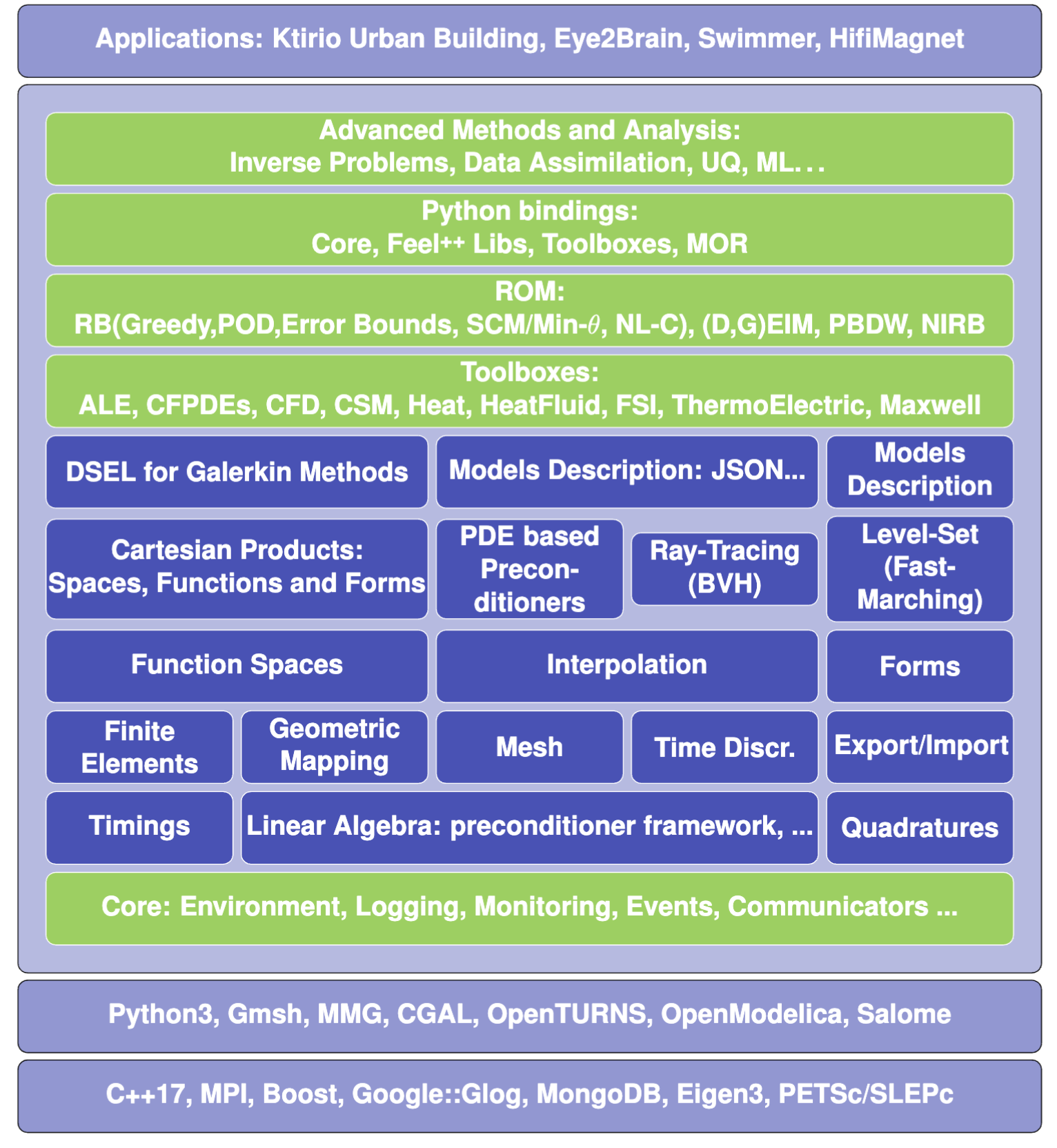
Applications
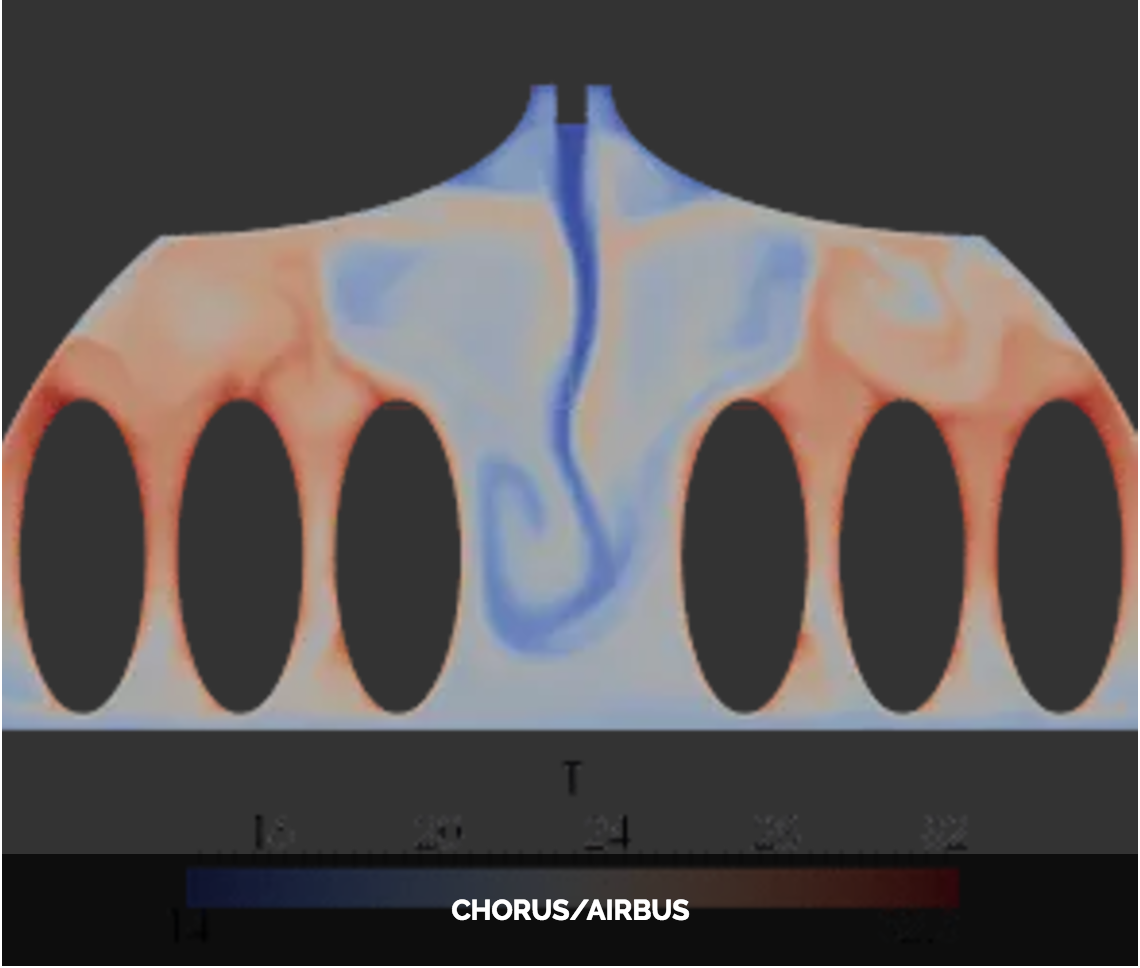



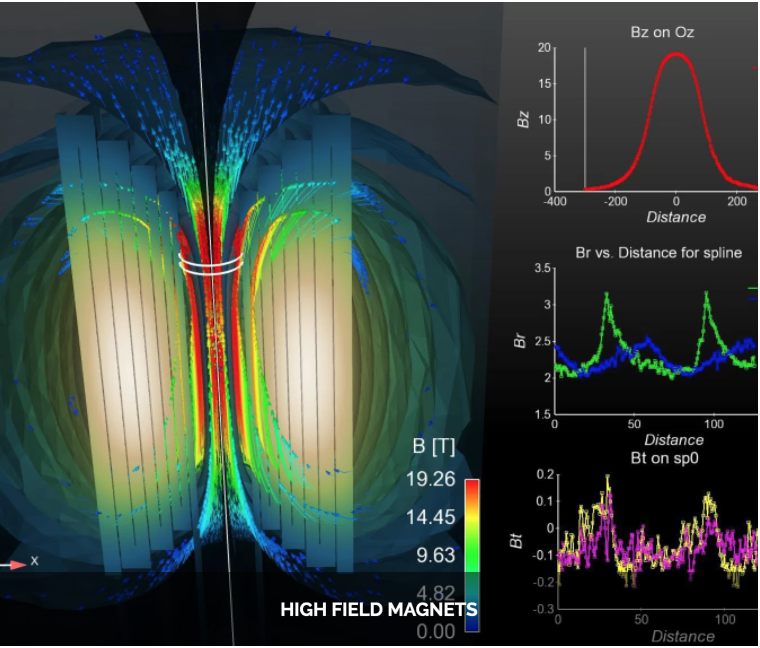


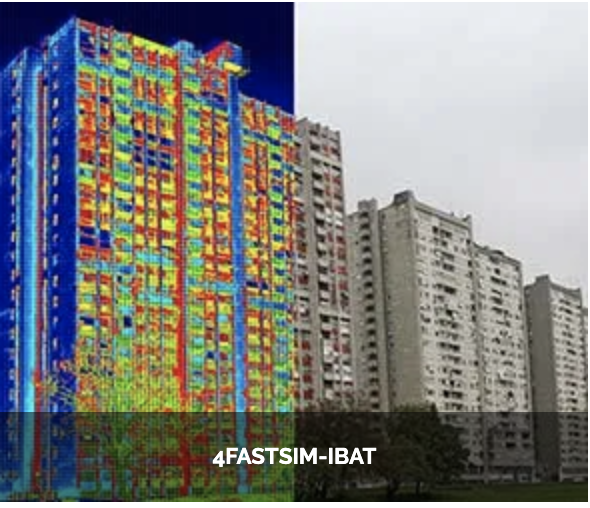
Feel++ Core Library
auto Vh = Pch<4>( mesh, markedelements(mesh, expr("<...>")) );
auto u = Vh->element(), v = Vh->element( g, "g" );
auto l = form1( _test = Vh );
l = integrate( _range = elements( support( Vh ) ),
_expr = f * id( v ) );
l += integrate( _range = markedfaces( support( Vh ), "Robin" ), _expr = -r_2 * id( v ) );
l += integrate( _range = markedfaces( support( Vh ), "Neumann" ), _expr = -un * id( v ) );
auto a = form2( _trial = Vh, _test = Vh );
a = integrate( _range = elements( support( Vh ) ),
_expr = inner( k * gradt( u ), grad( v ) ) );
a += integrate( _range = markedfaces( support( Vh ), "Robin" ), _expr = r_1*idt(u)*id(v));
a += on( _range = markedfaces( support(Vh), "Dirichlet" ), _rhs=l, _element=u, _expr = g );
a.solve( _rhs = l, _solution = u );A large range of numerical methods to solve PDEs: cG, dG, hdG, rb/mor, …
Finite Elements: \(L^2\), \(\mathbf{L}^2\), \(\mathbb{L}^2\), \(H^1\), \(\mathbf{H}^1\), \(\mathbb{H}^1\), \(\mathbf{H}(\text{div})\), \(\mathbf{H}(\text{curl})\), …
Value types: single, double, double double, quad double, mp precision and complex…
0D+t, 1D(+t), 2D(+t), 3D(+t)
DSEL for Galerkin methods in C++
Linear algebra: PETSc/SLEPc, Eigen, Boost::ublas …
Feel++ DSEL
// test strain tensor
auto def = 0.5*(grad(v) + trans(grad(v)));
// trial strain tensor
auto deft = 0.5*(gradt(u) + trans(gradt(u)));
// oseen
form2( _test=Xh, _trial=Xh, _matrix=M) =
// automatic quadature
integrate( elements(Xh->mesh()),
alpha*trans(idt(u))*id(v)
+ 2.0*nu*trace(trans(deft)*def)
+ trans(gradt(u)*idv(beta))*id(v)
- div(v)*idt(p) + divt(u)*id(q) );Advanced DSEL
Automatic differentiation
Automatic quadrature
Automatic assembly
Automatic parallelism
Feel++ Continuous Integration/Delivery/Deployment
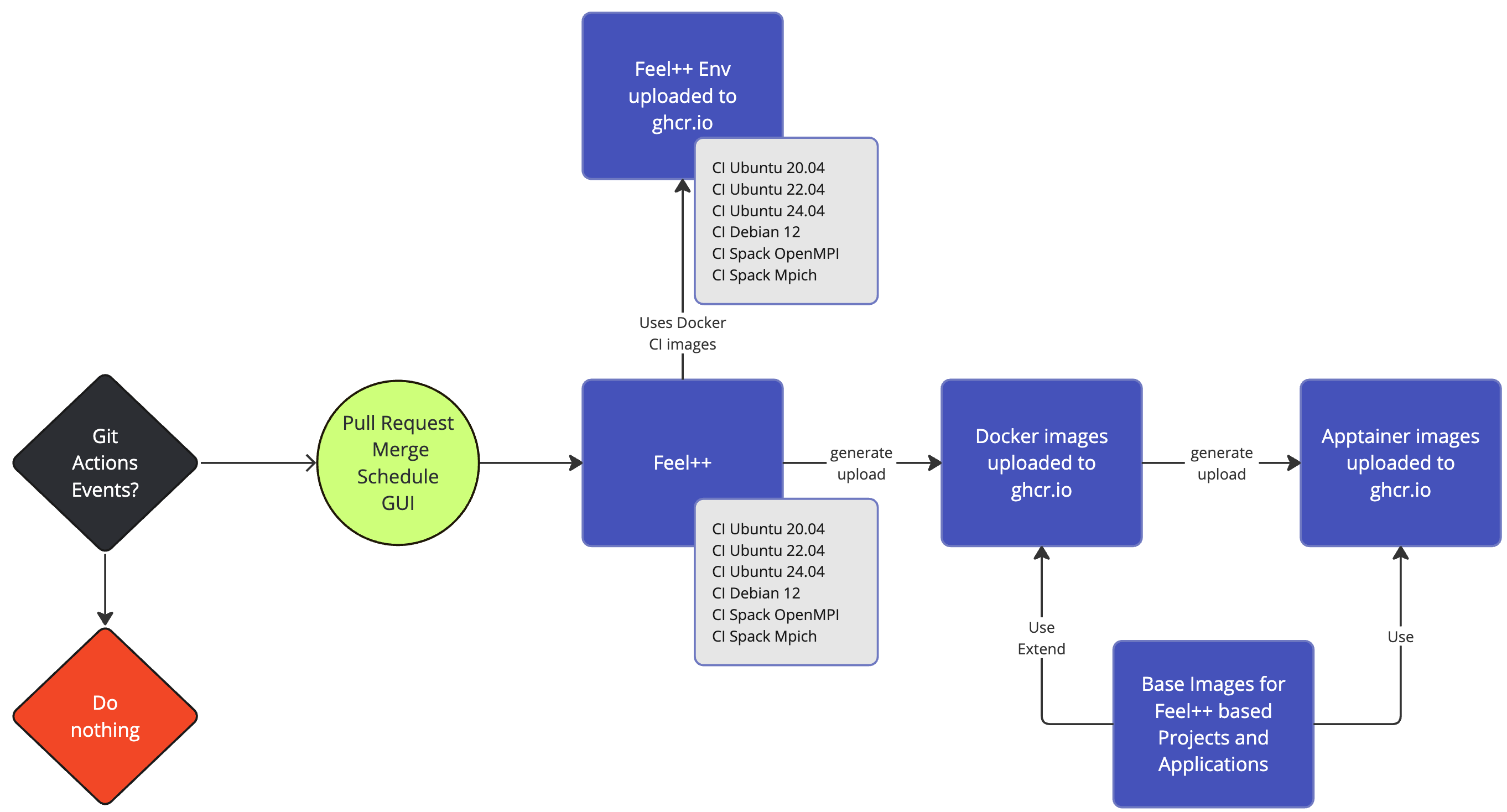
Feel++ Continuous Benchmarking
Feel++ Other features
// Named Arguments
auto Xh=Pch<1>(_mesh=mesh); (1)
auto u = Xh->element();
// expression handling
auto e3 = expr( "2*x*u*v:x:u:v" ); (2)
auto e3v = expr( e3, symbolExpr( "u", dxv( u ) ), symbolExpr( "v", Nx() ) );
// remeshing in seq and //
auto rm = remesh(_mesh=mesh,_params=<remeshing specs in json>); (3)
// Fast marching : compute distance to range
auto distToBoundary = distanceToRange( _space=Vh, _range=boundaryfaces( mesh ) ); (4)
auto distToBoundaryNarrowBand = (5)
distanceToRange( _space=Vh, _range=boundaryfaces( mesh ),
_max_distance=3.*mesh->hAverage() );| 1 | Expressivity: using our own named arguments library |
| 2 | Expressions handling: based on Ginac, handles automatic differentiation, JIT compilation |
| 3 | Remeshing in seq and parallel |
| 4 | Versatile levelset/fast marching framework |
| 5 | Narrow band FMM |
auto mesh = loadMesh<Mesh<Simplex<3,1>>();
auto Vh = Pch<2>( mesh ); (1)
auto u = Vh->element( inner(P()) ); (2)
auto e1 = expr( "u*u:u"); (3)
auto e2 = expr( "2*e1:e1");
auto e3base = expr( "3*e2:e2");
// handle composition
auto e3 = expr( e3base, symbolExpr("e1",e1), symbolExpr("e2",e2), symbolExpr("u",inner(P()) )); (4)
auto grad_e3 = grad<3>( e3 ); (5)
auto diff_e3_x_exact = 3*4*inner(P())*2*Px();
auto diff_e3_y_exact = 3*4*inner(P())*2*Py();
auto diff_e3_z_exact = 3*4*inner(P())*2*Pz();
auto grad_e3_exact = trans(vec(diff_e3_x_exact,diff_e3_y_exact,diff_e3_z_exact));
// compute error (machine error expected)
double error_grad_e3 = normL2(_range=elements(mesh),_expr= grad_e3 - grad_e3_exact ); (6)| 1 | create \(P^2_{c,h}\) |
| 2 | create an element of Vh such that \(u(x)=x^T x\) |
| 3 | create expressions |
| 4 | define symbols in expressions |
| 5 | differentiate the expressions |
| 6 | compute \(L^2\) error norm |
Feel++ Recent Developments:: Python
import feelpp
from feelpp.operators import *
mesh= feelpp.load(m, mesh_name, 0.1)
Xh = feelpp.functionSpace(mesh=mesh)
v=Xh.element()
v.on(range=feelpp.elements(mesh), expr=feelpp.expr("1"))
e_meas = mesh.measure()
M=mass(test=Xh,trial=Xh,range=feelpp.elements(mesh))
assert(abs(M.energy(v,v)-e_meas)<1e-10)
S=stiffness(test=Xh,trial=Xh,range=feelpp.elements(mesh))
assert(S.energy(v,v)<1e-10)
v.on(range=feelpp.elements(mesh), expr=feelpp.expr("x+y:x:y"))
assert(abs(S.energy(v,v)-2*e_meas)<1e-10)Parallel execution of toolboxes
Advanced multi-physics simulations
Advanced parametric studies including UQ
Reinforcement learning for micro-swimming
Ensemble Runs
Ensemble kalman filter in applications with sensors
import feelpp.core as fppc
from feelpp.toolboxes.core import *
from feelpp.toolboxes.fluid import *
from feelpp.toolboxes.heat import *
def simulate(toolbox, export=True, buildModelAlgebraicFactory=True,data=None):
toolbox.init(buildModelAlgebraicFactory)
#toolbox.printAndSaveInfo()
if toolbox.isStationary():
toolbox.solve()
if export:
toolbox.exportResults()
else:
if not toolbox.doRestart():
toolbox.exportResults(toolbox.timeInitial())
toolbox.startTimeStep()
while not toolbox.timeStepBase().isFinished():
if feelpp.Environment.isMasterRank():
print("time simulation: {}s/{}s with step: {}".format(toolbox.time(),toolbox.timeFinal(),toolbox.timeStep()))
toolbox.solve()
if export:
toolbox.exportResults()
toolbox.updateTimeStep()
return not toolbox.checkResults()
ht = heat(dim=3,order=1)
ok=simulate(ht)
meas = ht.postProcessMeasures().values()
df=pd.DataFrame([meas])
# do something with the pandas dataframe
....
cfd=fluid(dim=3,order=2)
ok=simulate(cfd)
....Toolboxes
CFD
Navier-Stokes incompressible 2D, 3D
Newtonian and non-newtonian
Multi-fluid support(levelset)
Moving domain support(ale)
Rigid and elastic body interaction.
Pressure BC.
Robust stab. methods.
WIP Turbulence Model.
Various formulations (e.g. Conservative, Curl, EMAC,…)
CSM
Linear elasticity
Large deformations, large displacements (Hyper elasticity)
Compressible, nearly incompressible materials
Multi-material support
Heat Transfer
2D and 3D heat transfer
High order in space and time
Diffusion and Convection
Robust Stab. Method
Thermo-Electric models including Seebeck/Peltier
Conjuguate heat transfer
RHT: on-going
Generic Toolbox :: Coefficient Form PDEs
Find scalar or vector fields \(u_i,i=1...,N\) such that
Coefficients can depend on the unknowns \(u_j,j=1 \ldots N\).
Automatic differentiation built on top of Feel++ expression handling
Various schemes based on Newton or Picard
"Models": {
"cfpdes":{ "equations":["equation1","equation2"] },
"equation1":{
"setup":{
"unknown":{"basis":"Pch1","name":"prey","symbol":"u"},
"coefficients":{
"c":"1", // diffusion
"a":"-( (1-equation1_u) - equation2_v/(equation1_u+thealpha) ):thealpha:equation1_u:equation2_v", // life and death
"d":"1" // time derivative
} } },
"equation2":{
"setup":{
"unknown":{ "basis":"Pch1", "name":"predator", "symbol":"v" },
"coefficients":{
"c":"thedelta:thedelta", // diffusion
"a":"-( (thebeta*equation1_u)/(equation1_u+thealpha) - thegamma ):thebeta:thealpha:thegamma:equation1_u", // life and death
"d":"1"// time derivative
} } } }
...Cahn Hilliard
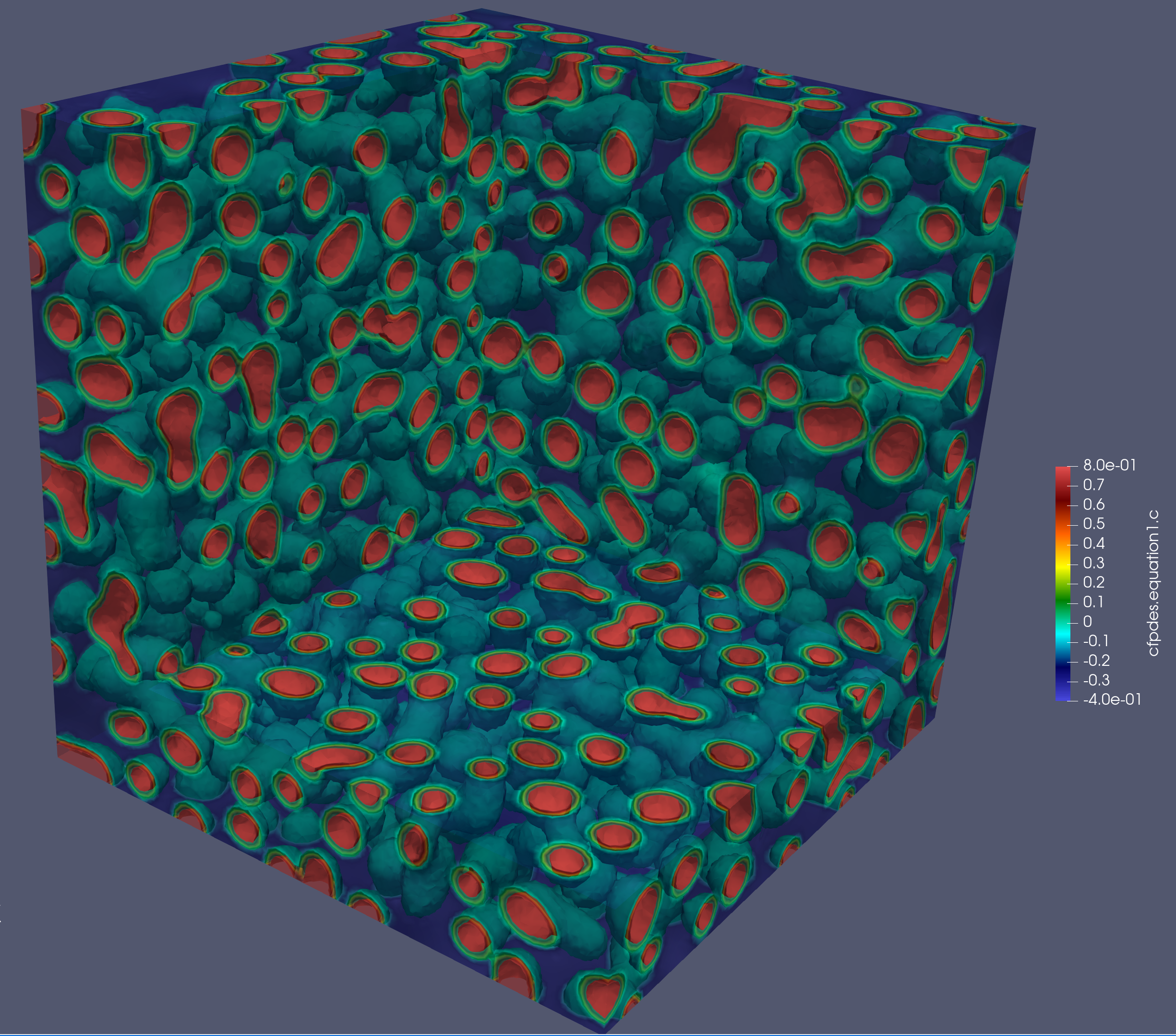
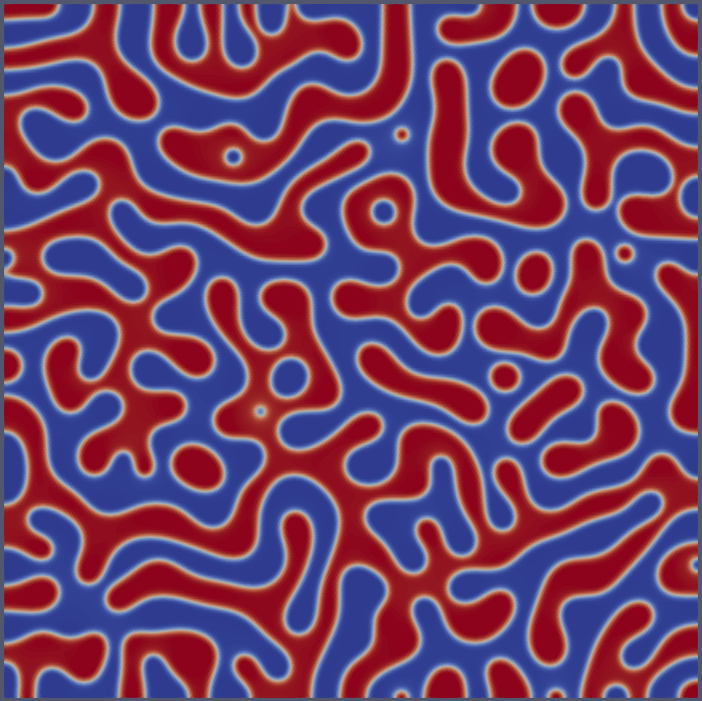
"Models":
{
"cfpdes":{ "equations":["equation1","equation2"] },
"equation1":{
"setup":{
"unknown":{"basis":"Pch1","name":"c","symbol":"c"},
"coefficients":{
"d": "1",
"gamma": "{-equation2_grad_mu_0,-equation2_grad_mu_1,-equation2_grad_mu_2}"
}}},
"equation2":{
"setup":{
"unknown":{"basis":"Pch1","name":"mu","symbol":"mu"},
"coefficients":{
"gamma":"{lambda*equation1_grad_c_0,lambda*equation1_grad_c_1, lambda*equation1_grad_c_2}",
"a":"1",
"f": "equation1_c^2*(equation1_c^2 - 1)"
} } }
}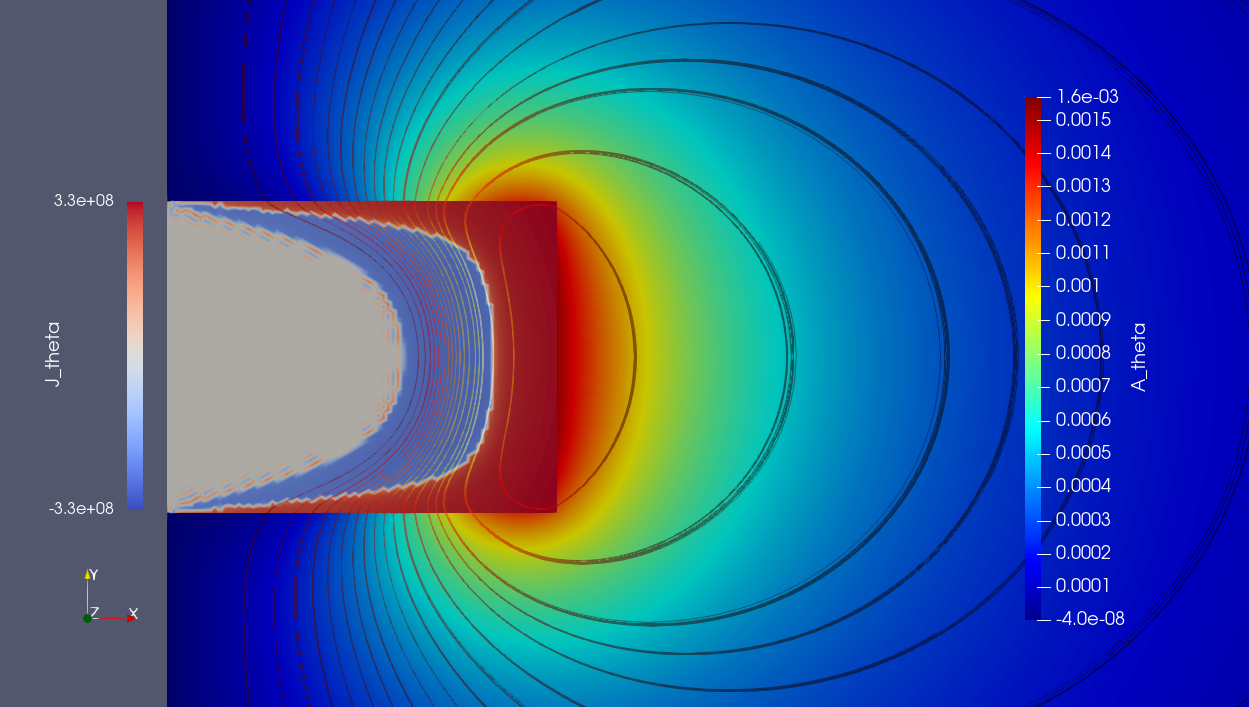
"Models":{
"cfpdes":{ "equations":"magnetic" },
"magnetic":{
"common":{"setup":{"unknown":{
"basis":"Pch1",
"name":"Atheta","
symbol":"Atheta"
}}},
"models":[{
"name":"magnetic_Conductor",
"materials":"Conductor",
"setup":{"coefficients":{
"c":"x/mu:x:mu",
"a":"1/mu/x:mu:x",
"d":"materials_Conductor_sigma*x
:materials_Conductor_sigma:x"}}
},{
"name":"magnetic_Air",
"materials":"Air",
"setup":{"coefficients":{
"c":"x/mu:x:mu",
"a":"1/mu/x:mu:x"}}}
]
}
}
...Toolbox:: Hybridized Discontinuous Galerkin
Second order elliptic problems
\(p\) is the potential
\(\mathbf{j}\) is the flux
\(\boldsymbol{u}: \Omega \rightarrow \mathbb{R}^{3}\) displacement
\(\underline{\boldsymbol{\epsilon}}(\boldsymbol{u}):=\frac{1}{2}\left(\nabla \boldsymbol{u}+(\nabla \boldsymbol{u})^{\top}\right)\) strain tensor
\(\underline{\boldsymbol{\sigma}}: \Omega \rightarrow S\) stress tensor where \(S\) is the set of all symmetric matrices in \(\mathbb{R}^{d\times d}\)
Methodology: HDG
Provides optimal approximation of both primal and flux/stress variables \(p/\boldsymbol{u}\) and \(\mathbf{j}/\underline{\boldsymbol{\sigma}}\) respectively;
Requires less globally coupled degrees of freedom than DG methods of comparable accuracy;
Allows local element-by-element postprocessing to obtain new approximations with enhanced accuracy and conservation properties
Some Applications
Electrostatic
Heat transfer
Flow in porous media
Elasticity and Poro-Elasticity
The integral boundary condition
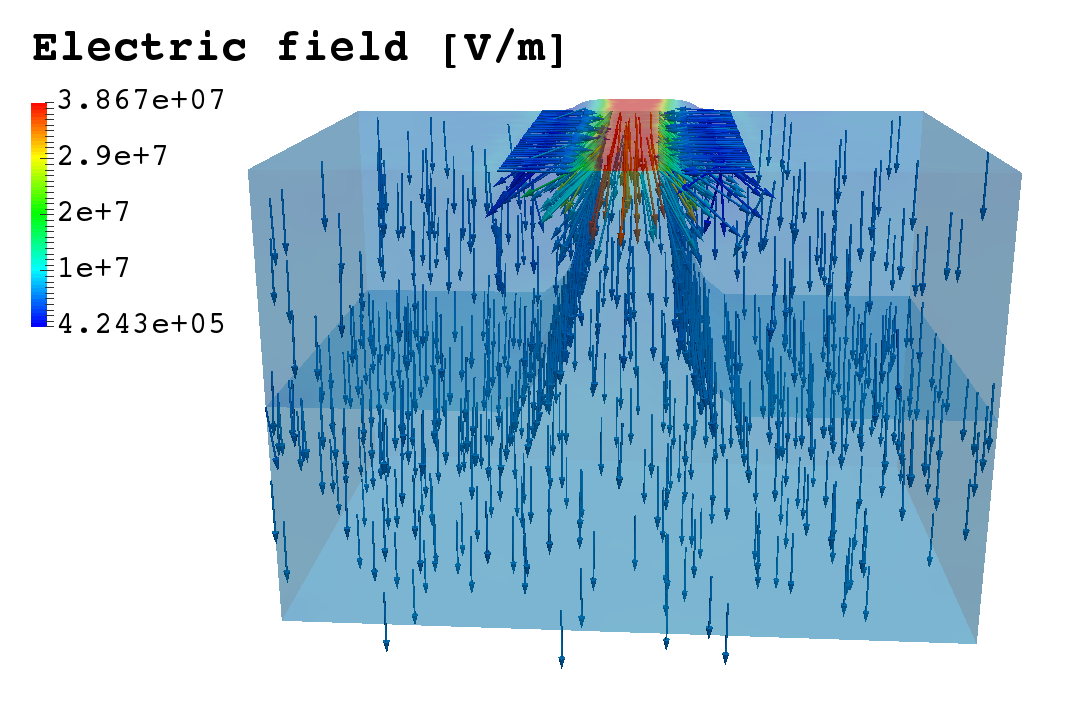
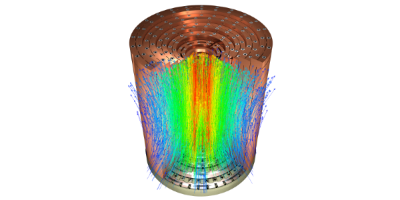
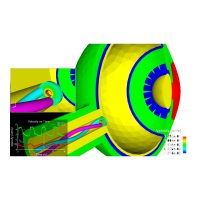
A HDG method for elliptic problems with integral boundary condition: Theory and Applications, Silvia Bertoluzza,Giovanna Guidoboni,Romain Hild,Daniele Prada,Christophe Prud’homme,Riccardo Sacco,Lorenzo Sala,Marcela Szopos, 2021 submitted
HDG Laplacian: formulation
\(\mathbf{T}_h\) the collection of elements \(K\) such that \(\Omega = \bigcup_{K\in \mathbf{T}_h} K\).
\(h:= max_{K \in \mathbf{T}_h} h_K\), \(\partial K\) the boundary of \(K\) with its measure \(|F|\)
\(\mathbf{n}_{\partial K}\) is the associated unit outward normal vector.
The skeleton of \(\mathbf{T}_h\) is the collection of all the faces of \(\mathbf{T}_h\) into the set \(\mathbf{F}_h\).
\(\mathbf{F}_h = \mathbf{F}_h^\Gamma \cup \mathbf{F}_h^0, \; \mathbf{F}_h^\Gamma = \mathbf{F}_h^D \cup \mathbf{F}_h^N \cup \mathbf{F}_h^{ibc}\)
\(V_h = \Pi_{K \in \mathbf{T}_h} V(K), \qquad V(K) = \left[ P_k (K) \right]^n\)
\(W_h = \Pi_{K\in\mathbf{T}_h} W(K), \qquad W(K) = \left[ P_k (K) \right]\)
\(\widetilde M_h = \{ \mu \in L^2(\mathbf{F}_h): \mu\rvert_F \in P_k(F) \; \forall F \in \mathbf{F}_h \setminus \mathbf{F}_h^{ibc} \},\)
\(M^*_h = \{ \mu \in C^0(\mathbf{F}^{ibc}_h): \mu\rvert_F \in P_0(F) \; \forall F \in \mathbf{F}_h^{ibc} \} \cong \mathbb{R}\)
\(M_h=\widetilde M_h \oplus M^*_h\)
HDG Laplacian: formulation
Discrete formulation Find \(\boldsymbol{j}_h \in V_h, \; p_h \in W_h\) and \(\hat{p}_h \in M_h\) such that:
HDG Laplacian
auto mesh=loadMesh(_mesh=new Mesh<Simplex<3>>); (1)
auto complement_integral_bdy = complement(faces(mesh), (2)
[&mesh]( auto const& e ) {
if ( e.hasMarker() &&
e.marker().matches(mesh->markerName("Ibc*") ) )
return true;
return false;
});
auto face_mesh = createSubmesh( mesh, complement_integral_bdy); (3)
auto ibc_mesh = createSubmesh( mesh, markedfaces(mesh,"Ibc*")); (4)| 1 | load mesh |
| 2 | build set of non ibc facets |
| 3 | \(\mathbf{F}_h^{ibc}\) and |
| 4 | \(\mathbf{F}_h\setminus\mathbf{F}_h^{ibc}\). |
Vh_ptr_t Vh = Pdhv<OrderP>( mesh); (1)
Wh_ptr_t Wh = Pdh<OrderP>( mesh );
Mh_ptr_t Mh = Pdh<OrderP>( face_mesh );
// only one degree of freedom
Ch_ptr_t Ch = Pch<0>(ibc_mesh );
// $n$ IBC
auto ibcSpaces = product( nb_ibc, Ch); (2)
auto Xh = product( Vh, Wh, Mh. ibcSpaces ); (3)| 1 | create the spaces \(V_h,W_h,\tilde{M}_h\) and \(M_h^*\). |
| 2 | handle arbirary number of IBCs |
| 3 | initialize spaces and product space |
HDG laplacian
auto a = blockform2( Xh )
auto rhs = blockform1( Xh );
. . .
// Assembling the right hand side
rhs(1_c) += integrate(_range=elements(mesh),_expr=-f*id(w));
. . .
// Assembling the main matrix
a(0_c,0_c) += integrate(_range=elements(mesh),
_expr=(trans(lambda*idt(u))*id(v)) );
. . .
//$\langle \hat{p}_h\rvert_{\tilde{M}_h}, \boldsymbol{v}_h^K \cdot {\boldsymbol{n}}_{\partial K}\rangle$ $$
a(0_c,2_c) += integrate(_range=internalfaces(mesh),
_expr=( idt(phat)*(leftface(trans(id(v))*N())+
rightface(trans(id(v))*N()))));a( 3_c, 0_c, i, 0 ) +=
integrate( _range=markedfaces(mesh,"Ibc"),
_expr=(trans(idt(u))*N()) * id(nu) );
auto U = Xh.element();
a.solve(_solution=U, _rhs=rhs, _name="hdg");
auto up = U(0_c);
auto pp = U(1_c);
auto phat = U(2_c);
auto ip = U(3_c,0);
// postprocessing
auto Whp = Pdh<OrderP+1>( mesh );
auto pps = product( Whp );
auto PP = pps.element();
auto ppp = PP(0_c);
auto b = blockform2( pps, solve::strategy::local, backend() );
b( 0_c, 0_c ) = integrate( _range=elements(mesh), _expr=inner(gradt(ppp),grad(ppp)));
auto ell = blockform1( pps, solve::strategy::local, backend() );
ell(0_c) = integrate( _range=elements(mesh), _expr=-lambda*grad(ppp)*idv(up));
b.solve( _solution=PP, _rhs=ell, _name="sc.post", _local=true);
ppp=PP(0_c);
ppp += -ppp.ewiseMean(P0dh)+pp.ewiseMean(P0dh);Toolbox HDG
Similar to CFPDEs, except that only one equation for now
time dependence
Other terms in the PDEs
non-linear coefficients
IBCs
arbitrary number
Coupling with 0D+t models using FMU
time splitting approach to avoid iterating
Extended to PoroElasticity
WIP: HHO support
WIP: Order reduction (RB and NiRB)
Future: merge with CFPDEs

Final Words
Develop Feel++ as a service
Streamline complex workflows for advanced usage/studies
data assimilation,
machine learning (interface with Scimba and Scikit-learn),
UQ on complex models (interface with OpenTURNs)
…
Go Digital twins
Building energy modeling (Ktirio Urban Building - CoE Hidalgo2)
High field magnets (LNCMI)
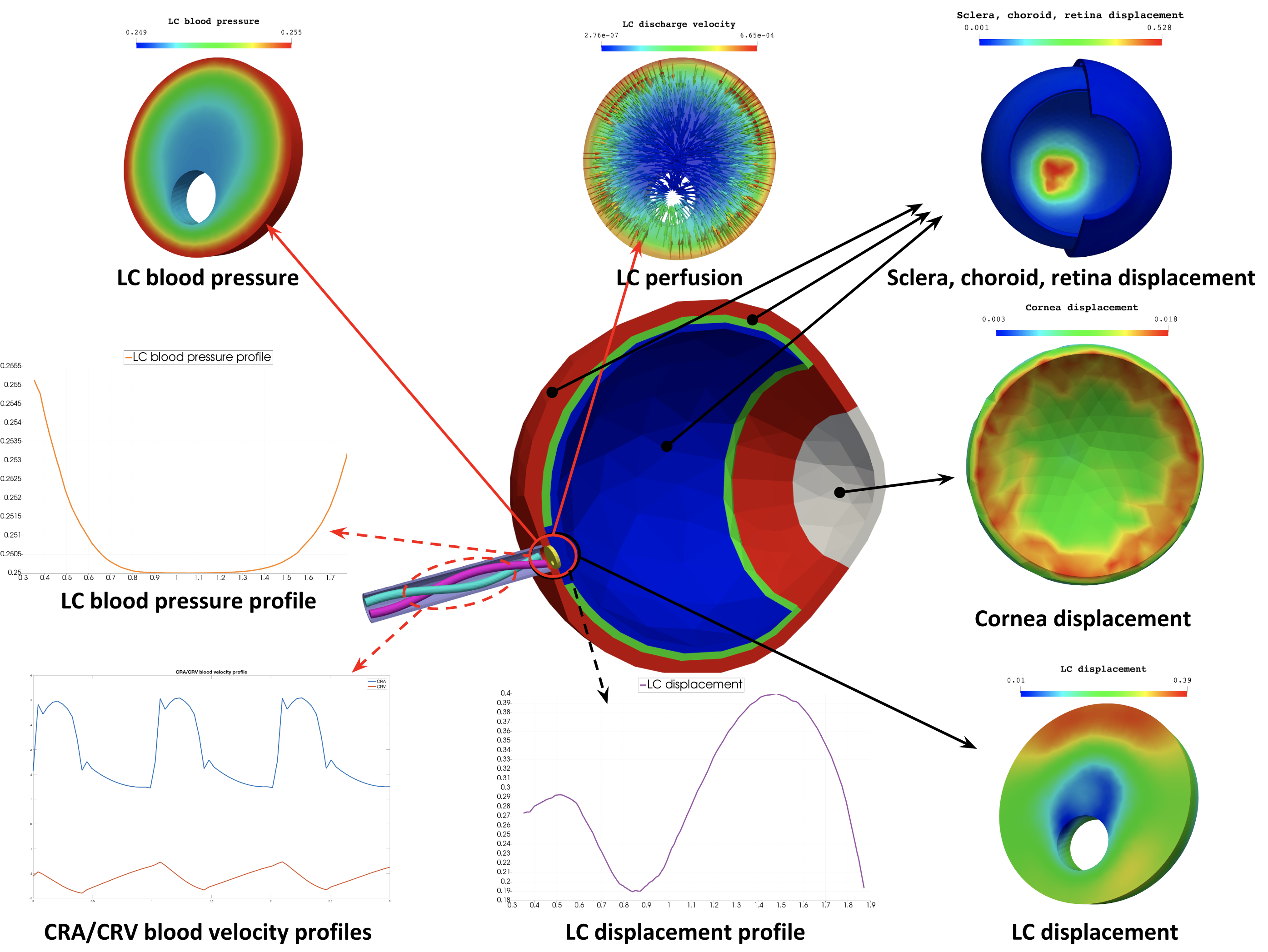
Eye (Eye2Brain)